Dataset: Wnt4 KO in Developing mouse kidney
Sarah Williams
Last updated: 2024-09-19
Checks: 7 0
Knit directory: spatialsnippets/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231017) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 511594f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/e_neighbourcellchanges.nb.html
Ignored: analysis/glossary.nb.html
Ignored: renv/library/
Ignored: renv/staging/
Unstaged changes:
Modified: .gitignore
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/d_Wnt4KO.Rmd) and HTML
(docs/d_Wnt4KO.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 511594f | swbioinf | 2024-09-19 | wflow_publish("analysis/") |
| html | 1549b68 | swbioinf | 2024-09-17 | Build site. |
| Rmd | 42d8712 | swbioinf | 2024-09-17 | wflow_publish("analysis/d_Wnt4KO.Rmd") |
| html | 2271297 | swbioinf | 2024-09-11 | Build site. |
| Rmd | 528f6cd | swbioinf | 2024-09-11 | wflow_publish("analysis/") |
Data from paper Forming nephrons promote nephron progenitor maintenance and branching morphogenesis via paracrine BMP4 signalling under the control of Wnt4 (Moreau et al. 2023)
This study included 10X chromium single cell RNAseq data from 4 conditions, with 3-4 E14.5 mice pooled per group.
- Sample1 (Wnt4FloxKO): Wnt4Flox/Flox Six2-Conditional Wnt4 Knockout
- Sample2 (Wnt4FloxHet): Wnt4Flox/+ Six2-Conditional Wnt4 Het
- Sample3 (Wnt4Het): Wnt4 GCE/+ Control Wnt4 Het
- Sample4 (Wnt4KO): Wnt4 GCE/GCE Knockout Wnt4
In that paper they explain that complete or conditional homozygous knockout of Wnt4 gene results in abnormal kidney development, and they use scRNAseq data to explore effects at cellular level. (Moreau et al. 2023)
Libraries
library(Seurat)Loading required package: SeuratObjectLoading required package: sp
Attaching package: 'SeuratObject'The following objects are masked from 'package:base':
intersect, t#library(SeuratObject)
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.0 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2 ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsData load
Loading data from a previously annotated object used in the analysis.
dataset_dir <- '~/projects/spatialsnippets/datasets'
project_data_dir <- file.path(dataset_dir,'Wnt4KO_Moreau2023')
seurat_file_loaded <- file.path(project_data_dir, "Wnt4KOE14.5_10_withdeconclust.rds")
seurat_file_saved <- file.path(project_data_dir, "Wnt4KOE14.5_11_ss.rds")Load and tidy data
so <- readRDS(seurat_file_loaded)
# There's alot of historical/working feilds in the metadata.
# Slim down to just useful ones.
keep_cols <- c(
"barcode","orig.ident","nCount_RNA","nFeature_RNA" ,"sample", # basic annotation/stats.
"Genotype","GTeffect", "GTshort" , # same info, 3 ways
"MT_pc", "cc" ,"G1" ,"S" ,"G2M", "doublet_score" , # was filtered on scrublet.
# "cluster_content","cluster_lab" # More detailed groupings used in working
"Celltype", "CelltypeCode", # final types in paper
"CelltypeBroad", "CelltypeBroadCode" # broader groupings
)
so@meta.data <- so@meta.data[,keep_cols]Data tour
DimPlot(so, group.by="CelltypeCode",label = TRUE) + NoLegend()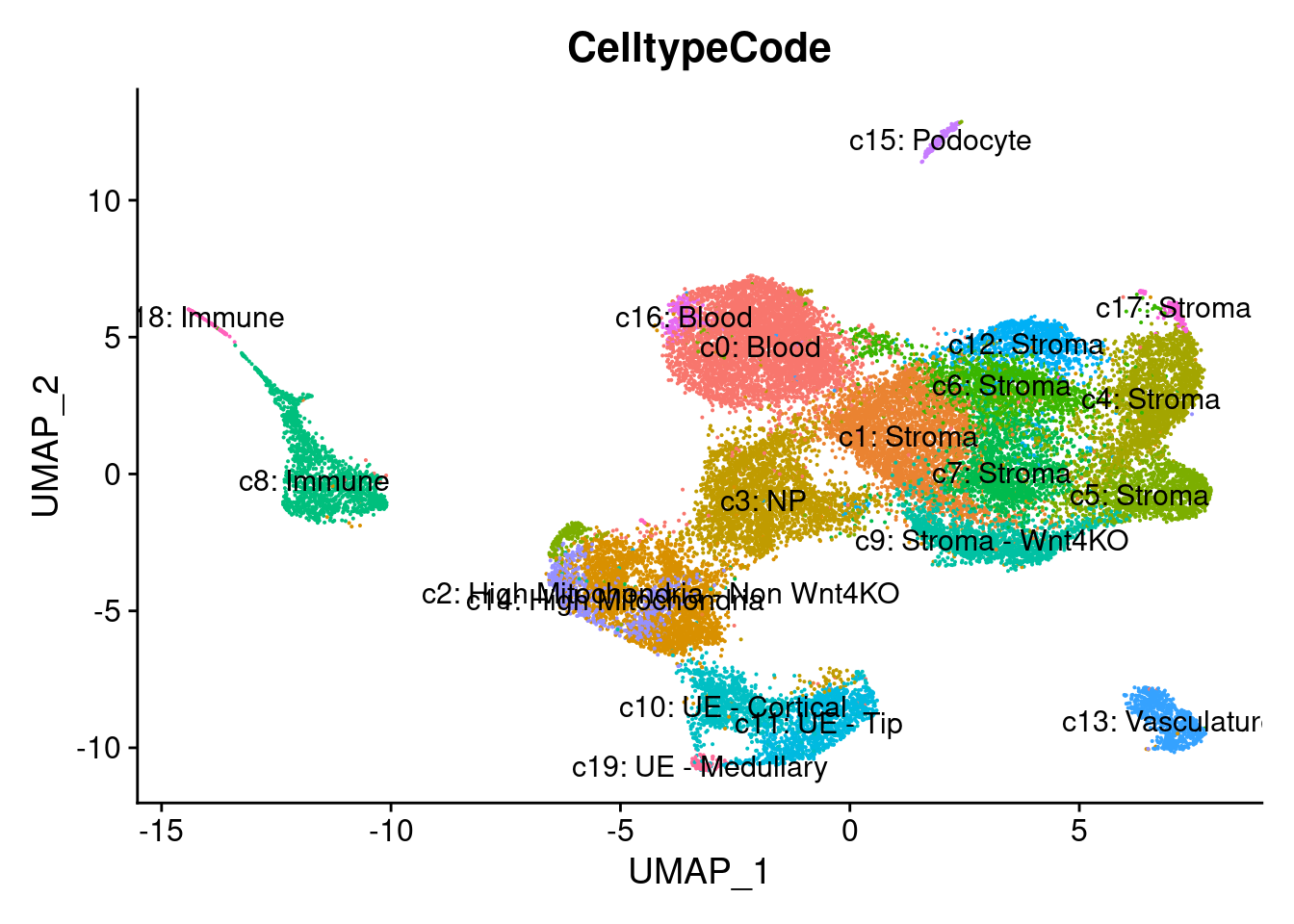
DimPlot(so, group.by="CelltypeBroad",label = TRUE) + NoLegend()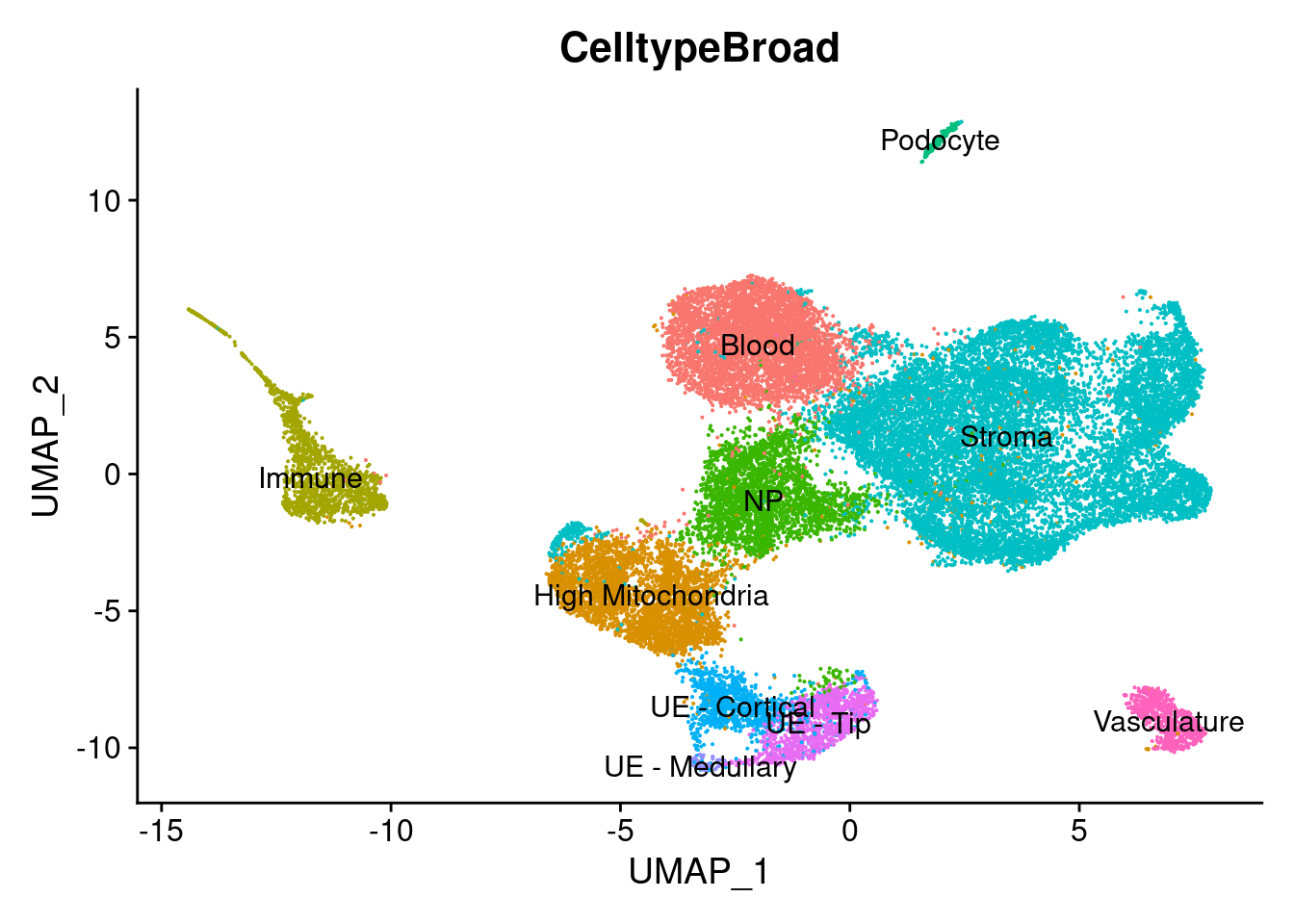
DimPlot(so, group.by="GTshort")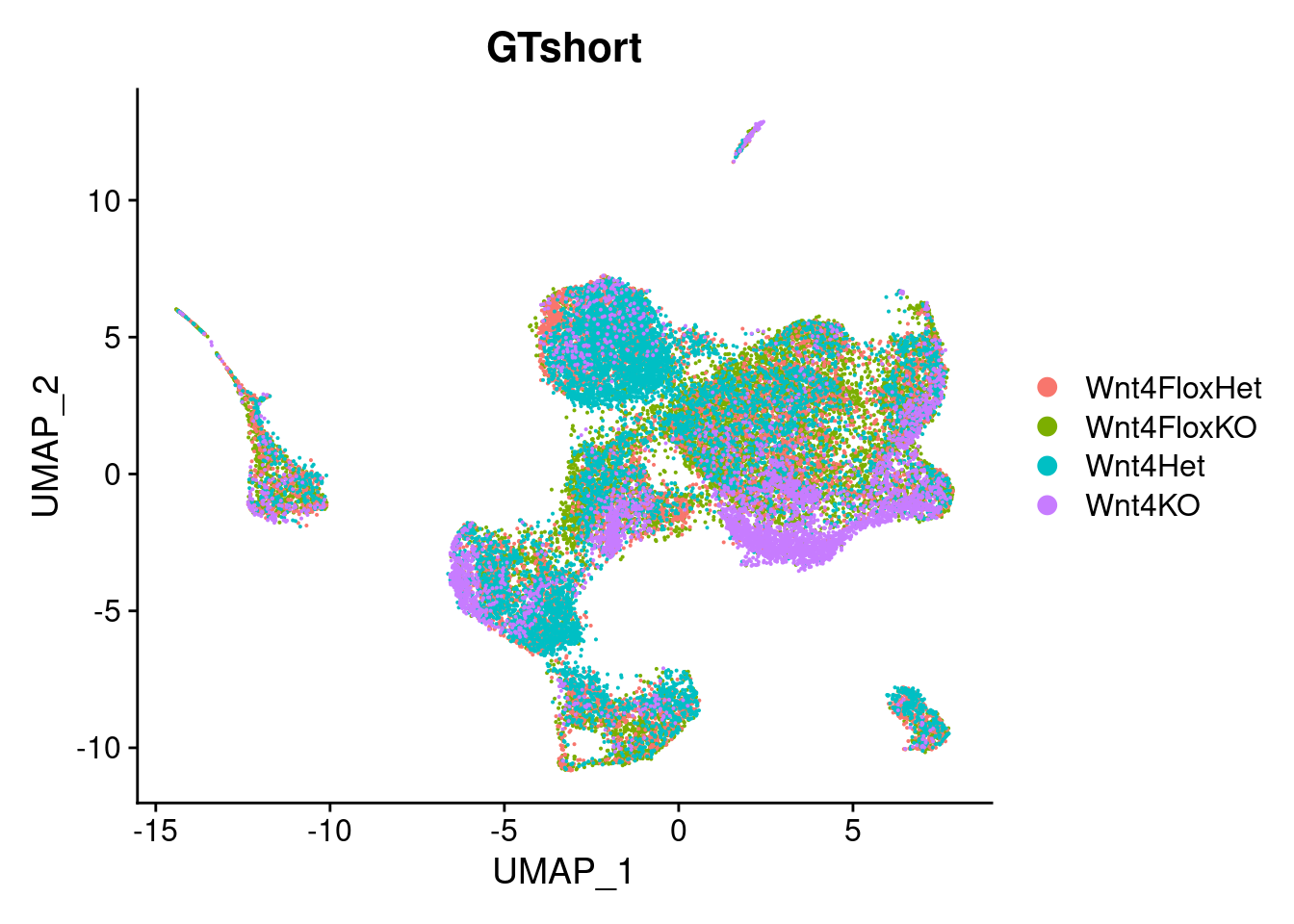
DimPlot(so, group.by="cc")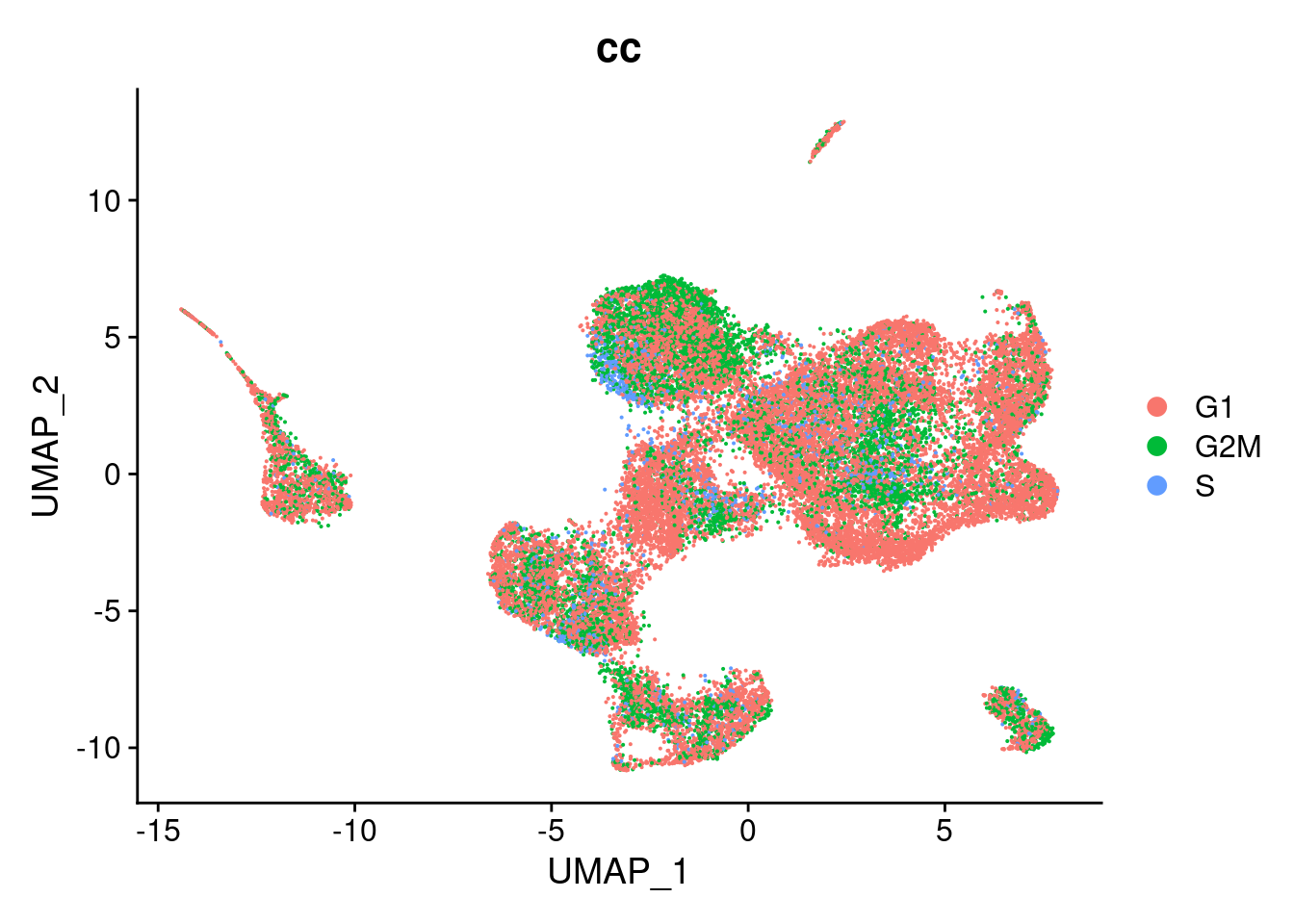
FeaturePlot(so, c('nCount_RNA','MT_pc'))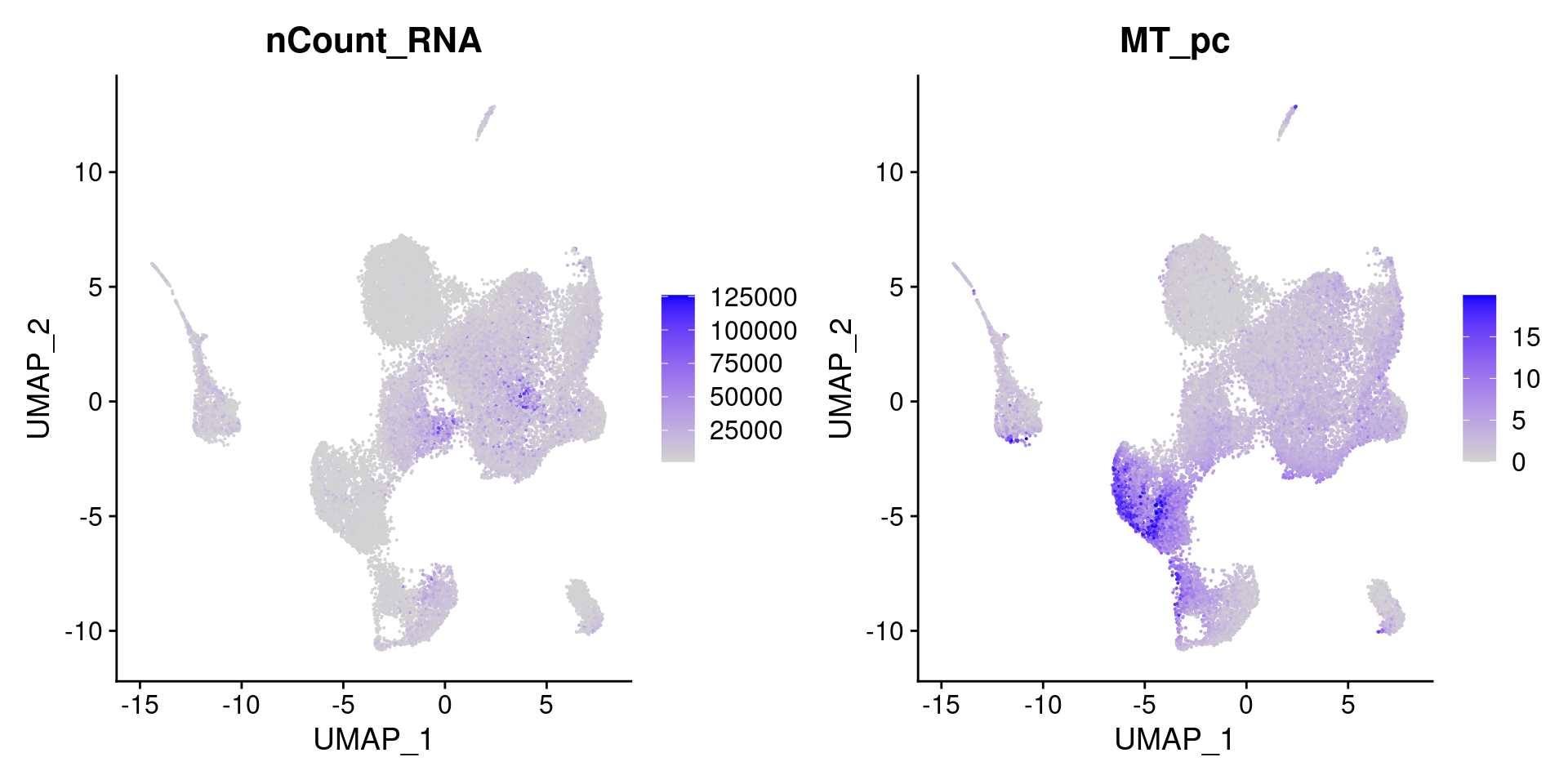
4 Samples, n=1 per group. Can use ‘GTshort’ to identify the sample.
select(so@meta.data, sample, Genotype, GTeffect, GTshort) %>%
as_tibble() %>%
group_by( sample, Genotype, GTeffect, GTshort) %>%
summarise(num_cells=n(), .groups = 'drop') %>%
DT::datatable()Save it
saveRDS(so, seurat_file_saved)References
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
[5] purrr_1.0.2 readr_2.1.5 tidyr_1.3.1 tibble_3.2.1
[9] ggplot2_3.5.0 tidyverse_2.0.0 Seurat_5.1.0 SeuratObject_5.0.2
[13] sp_2.1-3 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RColorBrewer_1.1-3 rstudioapi_0.16.0 jsonlite_1.8.8
[4] magrittr_2.0.3 spatstat.utils_3.0-4 farver_2.1.1
[7] rmarkdown_2.26 fs_1.6.3 vctrs_0.6.5
[10] ROCR_1.0-11 spatstat.explore_3.2-7 htmltools_0.5.8.1
[13] sass_0.4.9 sctransform_0.4.1 parallelly_1.37.1
[16] KernSmooth_2.23-22 bslib_0.7.0 htmlwidgets_1.6.4
[19] ica_1.0-3 plyr_1.8.9 plotly_4.10.4
[22] zoo_1.8-12 cachem_1.0.8 whisker_0.4.1
[25] igraph_2.0.3 mime_0.12 lifecycle_1.0.4
[28] pkgconfig_2.0.3 Matrix_1.6-5 R6_2.5.1
[31] fastmap_1.1.1 fitdistrplus_1.1-11 future_1.33.2
[34] shiny_1.8.1.1 digest_0.6.35 colorspace_2.1-0
[37] patchwork_1.2.0 ps_1.7.6 rprojroot_2.0.4
[40] tensor_1.5 RSpectra_0.16-1 irlba_2.3.5.1
[43] crosstalk_1.2.1 labeling_0.4.3 progressr_0.14.0
[46] timechange_0.3.0 fansi_1.0.6 spatstat.sparse_3.0-3
[49] httr_1.4.7 polyclip_1.10-6 abind_1.4-5
[52] compiler_4.3.2 withr_3.0.0 fastDummies_1.7.3
[55] highr_0.10 MASS_7.3-60.0.1 tools_4.3.2
[58] lmtest_0.9-40 httpuv_1.6.15 future.apply_1.11.2
[61] goftest_1.2-3 glue_1.7.0 callr_3.7.6
[64] nlme_3.1-164 promises_1.2.1 grid_4.3.2
[67] Rtsne_0.17 getPass_0.2-4 cluster_2.1.6
[70] reshape2_1.4.4 generics_0.1.3 gtable_0.3.4
[73] spatstat.data_3.0-4 tzdb_0.4.0 hms_1.1.3
[76] data.table_1.15.4 utf8_1.2.4 spatstat.geom_3.2-9
[79] RcppAnnoy_0.0.22 ggrepel_0.9.5 RANN_2.6.1
[82] pillar_1.9.0 spam_2.10-0 RcppHNSW_0.6.0
[85] later_1.3.2 splines_4.3.2 lattice_0.22-6
[88] renv_1.0.5 survival_3.5-8 deldir_2.0-4
[91] tidyselect_1.2.1 miniUI_0.1.1.1 pbapply_1.7-2
[94] knitr_1.45 git2r_0.33.0 gridExtra_2.3
[97] scattermore_1.2 xfun_0.43 matrixStats_1.2.0
[100] DT_0.33 stringi_1.8.3 lazyeval_0.2.2
[103] yaml_2.3.8 evaluate_0.23 codetools_0.2-20
[106] BiocManager_1.30.22 cli_3.6.2 uwot_0.1.16
[109] xtable_1.8-4 reticulate_1.35.0 munsell_0.5.1
[112] processx_3.8.4 jquerylib_0.1.4 Rcpp_1.0.12
[115] globals_0.16.3 spatstat.random_3.2-3 png_0.1-8
[118] parallel_4.3.2 dotCall64_1.1-1 listenv_0.9.1
[121] viridisLite_0.4.2 scales_1.3.0 ggridges_0.5.6
[124] leiden_0.4.3.1 rlang_1.1.3 cowplot_1.1.3