8 Cell cycle Assignment
In some datasets, the phase of cell cycle that a cell is in (G1/G2M/S) can account for alot of the observed transcriptomic variation. There may be clustering by phase, or separation in the UMAP by phase.
Seurat provides a simple method for assigning cell cycle state to each cell. Other methods are available.
More information about assigning cell cycle states to cells is in the cell cycle vignette
# A list of cell cycle markers, from Tirosh et al, 2015, is loaded with Seurat. We can
# segregate this list into markers of G2/M phase and markers of S phase
s.genes <- cc.genes$s.genes
g2m.genes <- cc.genes$g2m.genes
# Use those lists with the cell cycle scoring function in Seurat.
pbmc <- CellCycleScoring(pbmc, s.features = s.genes, g2m.features = g2m.genes)
#> Warning: The following features are not present in the
#> object: DTL, UHRF1, MLF1IP, EXO1, CASP8AP2, BRIP1, E2F8,
#> not searching for symbol synonyms
#> Warning: The following features are not present in the
#> object: FAM64A, BUB1, HJURP, CDCA3, TTK, CDC25C, DLGAP5,
#> CDCA2, ANLN, GAS2L3, not searching for symbol synonymsWhich adds S.Score, G2M.Score and Phase calls to the metadata.
head(pbmc@meta.data)
#> orig.ident nCount_RNA nFeature_RNA
#> AAACATACAACCAC-1 pbmc3k 2419 779
#> AAACATTGAGCTAC-1 pbmc3k 4903 1352
#> AAACATTGATCAGC-1 pbmc3k 3147 1129
#> AAACCGTGCTTCCG-1 pbmc3k 2639 960
#> AAACCGTGTATGCG-1 pbmc3k 980 521
#> AAACGCACTGGTAC-1 pbmc3k 2163 781
#> percent.mt RNA_snn_res.0.5 seurat_clusters
#> AAACATACAACCAC-1 3.0177759 2 6
#> AAACATTGAGCTAC-1 3.7935958 3 1
#> AAACATTGATCAGC-1 0.8897363 2 0
#> AAACCGTGCTTCCG-1 1.7430845 1 5
#> AAACCGTGTATGCG-1 1.2244898 6 8
#> AAACGCACTGGTAC-1 1.6643551 2 0
#> RNA_snn_res.0.1 RNA_snn_res.0.2
#> AAACATACAACCAC-1 0 0
#> AAACATTGAGCTAC-1 3 3
#> AAACATTGATCAGC-1 0 0
#> AAACCGTGCTTCCG-1 1 1
#> AAACCGTGTATGCG-1 2 2
#> AAACGCACTGGTAC-1 0 0
#> RNA_snn_res.0.3 RNA_snn_res.0.4
#> AAACATACAACCAC-1 0 2
#> AAACATTGAGCTAC-1 3 3
#> AAACATTGATCAGC-1 0 2
#> AAACCGTGCTTCCG-1 1 1
#> AAACCGTGTATGCG-1 2 6
#> AAACGCACTGGTAC-1 0 2
#> RNA_snn_res.0.6 RNA_snn_res.0.7
#> AAACATACAACCAC-1 1 1
#> AAACATTGAGCTAC-1 3 3
#> AAACATTGATCAGC-1 1 1
#> AAACCGTGCTTCCG-1 2 2
#> AAACCGTGTATGCG-1 6 6
#> AAACGCACTGGTAC-1 1 1
#> RNA_snn_res.0.8 RNA_snn_res.0.9
#> AAACATACAACCAC-1 6 1
#> AAACATTGAGCTAC-1 2 2
#> AAACATTGATCAGC-1 1 1
#> AAACCGTGCTTCCG-1 4 4
#> AAACCGTGTATGCG-1 8 7
#> AAACGCACTGGTAC-1 1 1
#> RNA_snn_res.1 RNA_snn_res.1.1
#> AAACATACAACCAC-1 6 6
#> AAACATTGAGCTAC-1 2 2
#> AAACATTGATCAGC-1 1 1
#> AAACCGTGCTTCCG-1 4 4
#> AAACCGTGTATGCG-1 8 8
#> AAACGCACTGGTAC-1 1 1
#> RNA_snn_res.1.2 RNA_snn_res.1.3
#> AAACATACAACCAC-1 6 8
#> AAACATTGAGCTAC-1 2 2
#> AAACATTGATCAGC-1 1 0
#> AAACCGTGCTTCCG-1 4 5
#> AAACCGTGTATGCG-1 8 9
#> AAACGCACTGGTAC-1 1 0
#> RNA_snn_res.1.4 RNA_snn_res.1.5
#> AAACATACAACCAC-1 8 9
#> AAACATTGAGCTAC-1 2 2
#> AAACATTGATCAGC-1 0 1
#> AAACCGTGCTTCCG-1 5 4
#> AAACCGTGTATGCG-1 9 8
#> AAACGCACTGGTAC-1 0 1
#> RNA_snn_res.1.6 RNA_snn_res.1.7
#> AAACATACAACCAC-1 8 8
#> AAACATTGAGCTAC-1 1 1
#> AAACATTGATCAGC-1 0 0
#> AAACCGTGCTTCCG-1 3 3
#> AAACCGTGTATGCG-1 7 7
#> AAACGCACTGGTAC-1 0 0
#> RNA_snn_res.1.8 RNA_snn_res.1.9
#> AAACATACAACCAC-1 7 6
#> AAACATTGAGCTAC-1 1 1
#> AAACATTGATCAGC-1 0 0
#> AAACCGTGCTTCCG-1 3 3
#> AAACCGTGTATGCG-1 8 8
#> AAACGCACTGGTAC-1 0 0
#> RNA_snn_res.2 Naive_CD4_T1 cell_label
#> AAACATACAACCAC-1 6 1.22824523 Naive_CD4_T
#> AAACATTGAGCTAC-1 1 -0.08111043 <NA>
#> AAACATTGATCAGC-1 0 -0.37682601 <NA>
#> AAACCGTGCTTCCG-1 5 -0.72739714 <NA>
#> AAACCGTGTATGCG-1 8 -1.17396454 <NA>
#> AAACGCACTGGTAC-1 0 -0.63807586 <NA>
#> cluster celltype pretty_cluster_labels
#> AAACATACAACCAC-1 c2 Memory CD4+ c2: Memory CD4+
#> AAACATTGAGCTAC-1 c3 B c3: B
#> AAACATTGATCAGC-1 c2 Memory CD4+ c2: Memory CD4+
#> AAACCGTGCTTCCG-1 c1 CD14+ Mono c1: CD14+ Mono
#> AAACCGTGTATGCG-1 c6 NK c6: NK
#> AAACGCACTGGTAC-1 c2 Memory CD4+ c2: Memory CD4+
#> SingleR.labels S.Score G2M.Score
#> AAACATACAACCAC-1 T_cells 0.09853841 -0.044716507
#> AAACATTGAGCTAC-1 B_cell -0.02364305 -0.046889929
#> AAACATTGATCAGC-1 T_cells -0.02177266 0.074841537
#> AAACCGTGCTTCCG-1 Monocyte 0.03794398 0.006575446
#> AAACCGTGTATGCG-1 NK_cell -0.03309970 0.027910063
#> AAACGCACTGGTAC-1 T_cells -0.04814181 -0.078164839
#> Phase
#> AAACATACAACCAC-1 S
#> AAACATTGAGCTAC-1 G1
#> AAACATTGATCAGC-1 G2M
#> AAACCGTGCTTCCG-1 S
#> AAACCGTGTATGCG-1 G2M
#> AAACGCACTGGTAC-1 G1We can then check the cell phase on the UMAP. In this dataset, phase isn’t driving the clustering, and would not require any further handling.
DimPlot(pbmc, reduction = 'umap', group.by = "Phase")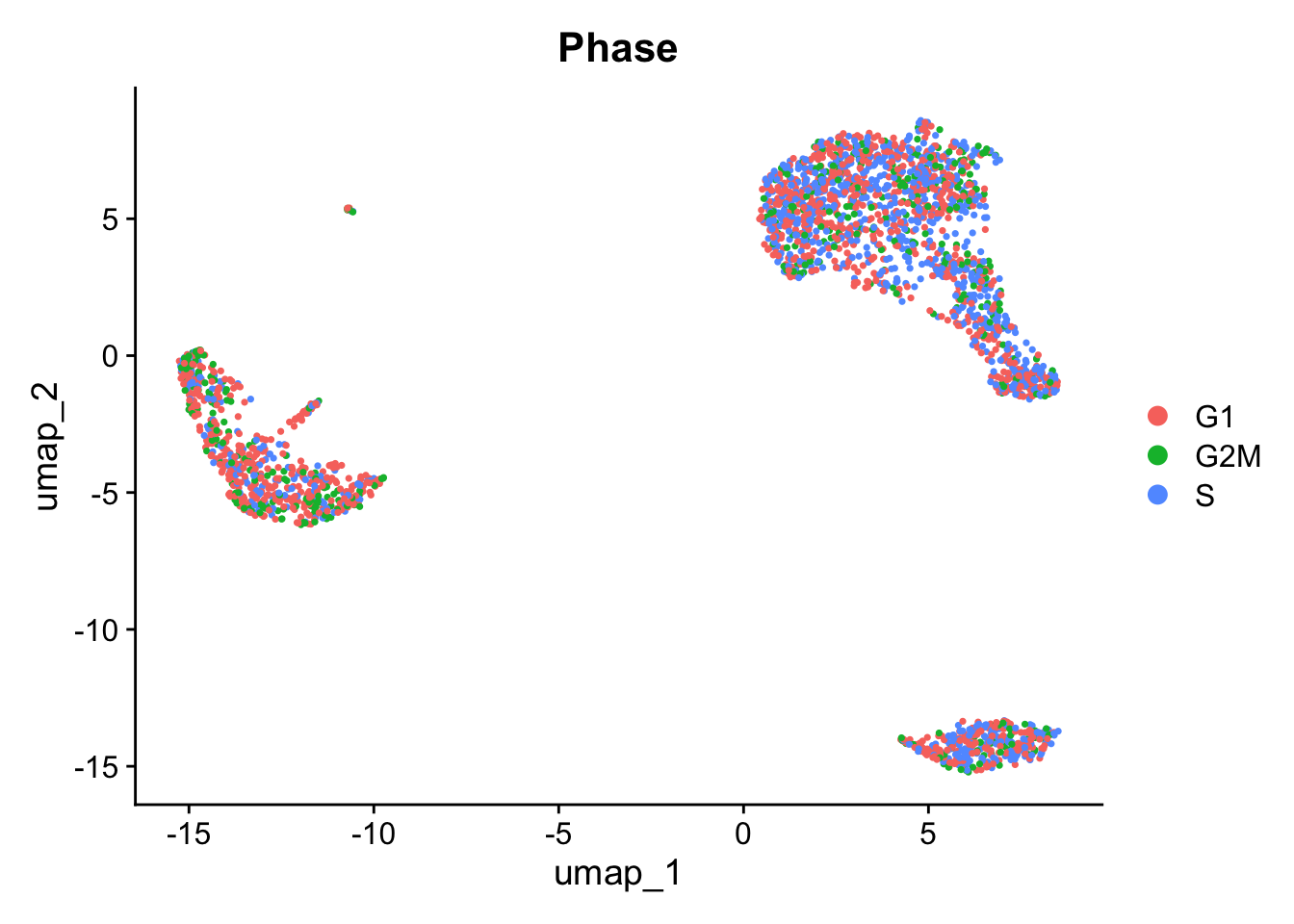
Where a bias is present, your course of action depends on the task at hand. It might involve ‘regressing out’ the cell cycle variation when scaling data ScaleData(kang, vars.to.regress="Phase"), omitting cell-cycle dominated clusters, or just accounting for it in your differential expression calculations.
If you are working with non-human data, you will need to convert these gene lists, or find new cell cycle associated genes in your species.